the most appealing proposals in this direction relies on kernel-based distances, expressed in terms of the embedding
transformation µPin (1); see [10].
The kernel kinvolved in this methodology depends, almost unavoidably, on some tuning parameter λ, typically a
scale factor. Therefore, we actually have a family of kernels, kλ, for λ∈Λ, where Λis usually a subset of Rk(k≥1).
For instance, the popular family of Gaussian kernels with parameter λ∈(0,∞)is defined by
kλ(x,y)=exp −λx−y2,for x,y∈X,(2)
where ⋅is a norm in X. Unfortunately, there is no general rule to know a priori which kernel works best with the
available data. In other words, the choice of λis, to some extent, arbitrary but not irrelevant, as it could remarkably
affect the final output. For example, very small or very large choices of λin (2) result in null discrepancies, which have
no ability to distinguish distributions. The selection of λis hence a delicate problem that has not been satisfactorily
solved so far. This is what we call the kernel trap: a bad choice of the parameter leading to poor results. Although
this problem was not explicitly considered in [10] and subsequent works on this topic, the authors were aware of this
relevant question; in practice, they use a heuristic choice of λ.
Further, a parameter-dependent method might be an obstacle for practitioners who are often reluctant to use
procedures depending on auxiliary, hard-to-interpret parameters. We thus find here a particular instance of the trade-
offbetween power and applicability: as stated in [47], the practical power of a statistical procedure is defined as “the
product of the mathematical power by the probability that the procedure will be used” (Tukey credits to Churchill
Eisenhart for this idea). From this perspective, our proposal can be viewed as an attempt to make kernel-based
homogeneity tests more usable by getting rid of the tuning parameter(s). Roughly speaking, the idea that we propose
to avoid selecting a specific value of λwithin the family {kλ∶λ∈Λ}is to take the supremum over the set of parameters
Λof the resulting family of kernel-distances. We call this approach the uniform kernel trick, as we map the data into
many functional spaces at the same time and use, as test statistic, the supremum of the corresponding kernel distances.
We believe that this methodology could be successfully applied as well in supervised classification, though this topic
is not considered in this work.
The topic of this paper
Two-sample tests, also called homogeneity tests, aim to decide whether or not it can be accepted that two random
elements have the same distribution, using the information provided by two independent samples. This problem is
omnipresent in practice on account of their applicability to a great variety of situations, ranging from biomedicine
to quality control. Since the classical Student’s t-tests or rank-based (Mann-Whitney, Wilcoxon, . . . ) procedures,
the subject has received an almost permanent attention from the statistical community. In this work we focus on
two-sample tests valid, under broad assumptions, for general settings in which the data are drawn from two random
elements Xand Ytaking values in a general space X. The set Xis the “sample space” or “feature space” in the
Machine Learning language. In the important particular case X=L2([0,1]),Xand Yare stochastic processes and
the two-sample problem lies within the framework of Functional Data Analysis (FDA).
Many important statistical methods, including goodness of fit and homogeneity tests, are based on an appropri-
ate metric (or discrepancy measure) that allows groups or distributions to be distinguished. Probability distances or
semi-distances reveal to the practitioner the dissimilarity between two random quantities. Therefore, the estimation of
a suitable distance helps detect significant differences between two populations. Some well-known, classic examples
of such metrics are the Kolmogorov distance, that leads to the popular Kolmogorov-Smirnov statistic, and L2-based
discrepancy measures, leading to Cram´
er-von Mises or Anderson-Darling statistics. These methods, based on cumu-
lative distribution functions, are no longer useful with high-dimensional or non-Euclidean data, as in FDA problems.
For this reason we follow a different strategy based on more adaptable metrics between general probability measures.
The energy distance (see the review by [41]) and the associated distance covariance, as well as kernel distance,
represent a step forward in this direction since they can be calculated with relative ease for high-dimensional distri-
butions. In [28] the relationships among these metrics in the context of hypothesis testing are discussed. In this paper
we consider an extension, as well as an alternative mathematical approach, for the two-sample test in [10]. These au-
thors show that kernel-based procedures perform better than other more classical approaches when dimension grows,
although they are strongly dependent on the choice of the kernel parameter.
2

 2024-12-12 388
2024-12-12 388
 2024-12-12 48
2024-12-12 48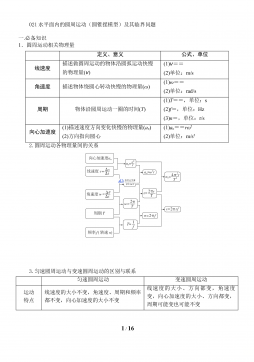
 2025-01-04 46
2025-01-04 46
 2025-04-08 7
2025-04-08 7
 2025-04-08 4
2025-04-08 4
 2025-04-08 4
2025-04-08 4
 2025-04-08 11
2025-04-08 11
 2025-04-08 13
2025-04-08 13
 2025-04-08 10
2025-04-08 10
 2025-04-08 11
2025-04-08 11






 渝公网安备50010702506394
渝公网安备50010702506394
